A gene co-expression network -- Dynamic Ca2+-dependent transcription links metabolic stress to impaired β-cell identity/p>
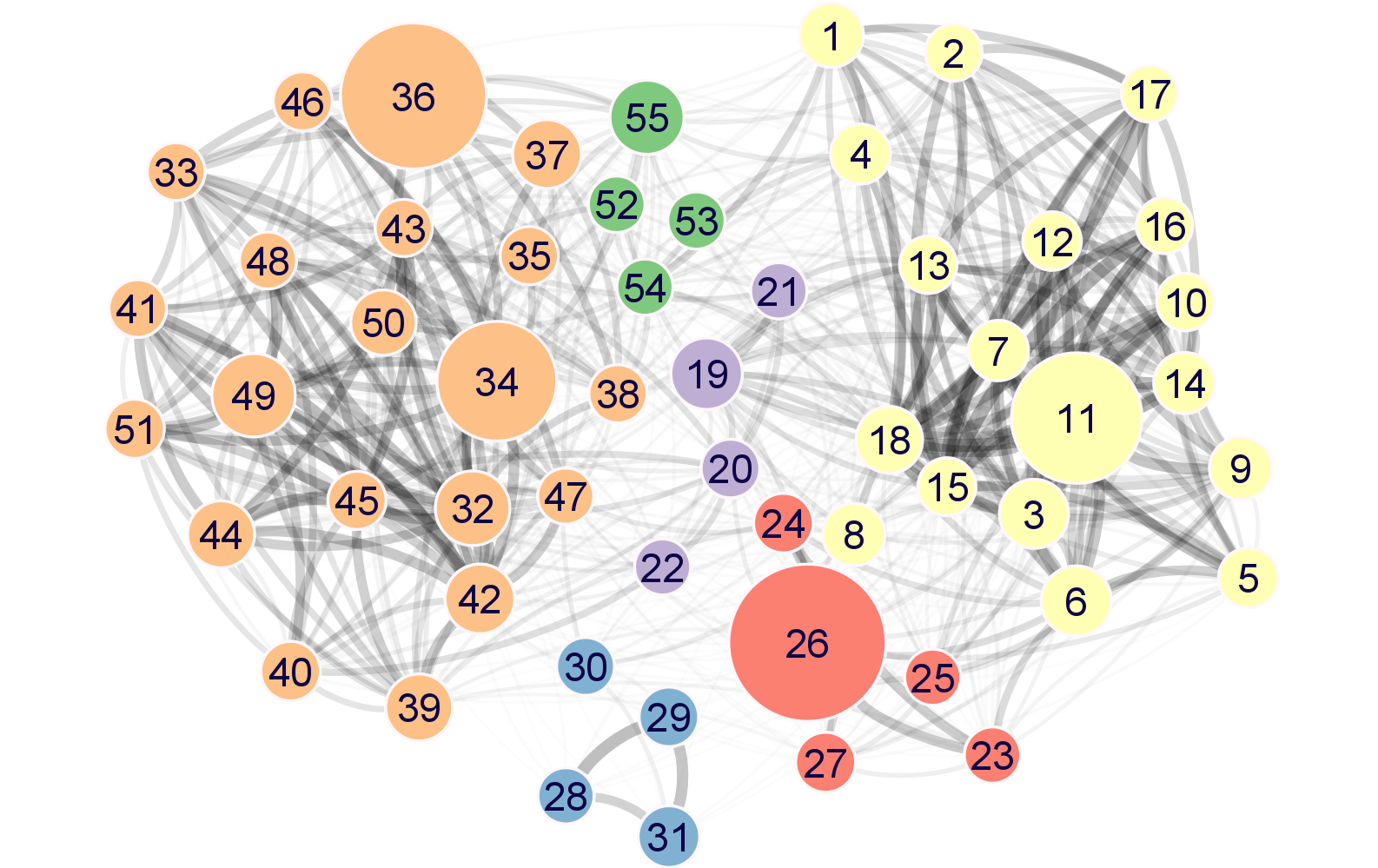
Each node in the meta-network represents a module of highly co-expressed genes. The meta-network is defined by correlations between module eigengenes and partitions modules into three distinct strongly connected module groups.
Please cite:
Anna B. Osipovich, et al. Dynamic Ca2+-dependent transcription links metabolic stress to impaired β-cell identity. In preparation.
For Biologists
We have prepared Cytoscape *session* files that you can download and simply open with Cytoscape to view and interact with the network meta-modules.
- Option 1 - Download Cytoscape file for all genes, then select the appropriate metamodule
- Option 2 - Download Cytoscape file for "TR" genes, filtered via transcription regulatory activity (GO:0140110)
For Bioinformaticians
For each meta-module, you will find below:
- Network image for "all genes" meta-modules
- Statistics about number of nodes and edges for "all genes" meta-modules
- Cytoscape JSON network file (compressed via ZSTD) for all genes
- Cytoscape JSON network file (compressed via ZSTD) for all genes identified as having transcription regulator activity (GO:0140110)
| Meta-module | Image | # nodes1 | # of edges2 | All genes | Transcription regulatory activity genes |
|---|---|---|---|---|---|
| A | 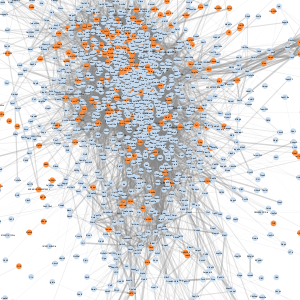 |
2,110 | 1,831,174 | Download | Download |
| B | 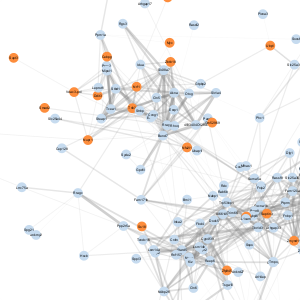 |
319 | 67,374 | Download | Download |
| C | 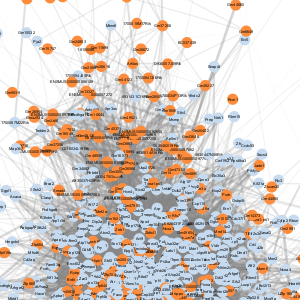 |
1,159 | 1,173,480 | Download | Download |
| D | 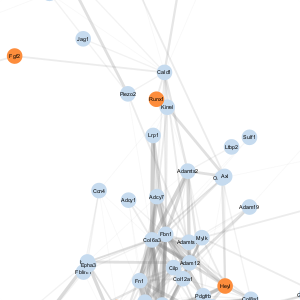 |
257 | 34,882 | Download | Download |
| E | 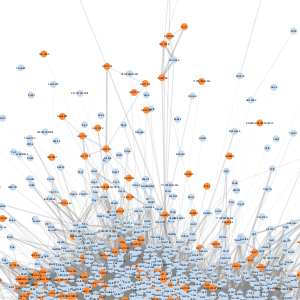 |
3,237 | 4,041,196 | Download | Download |
| F | 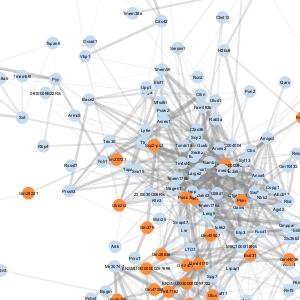 |
334 | 63,296 | Download | Download |
Legend
- A node represents a gene
- An edge represents a connection between two nodes and the number of edges shown is representative of edges with correlations of greater than 0.5
FAQ
Where can I get Cytoscape?
You can download it here.
How do I load network files in Cytoscape?
In order to load a network from a local file you can select File → Import → Network → File... Choose the correct file in the file chooser dialog and press Open. After you choose a network file, another dialog will pop up. Here, you can choose either to create a new network collection for the new network, or load the new network into an existing network collection. When you choose the latter, make sure to choose the right mapping column to map the new network to the existing network collection.
ZSTD? What's that?
Zstandard is a real-time compression algorithm, providing high compression ratios. It's like ZIP, but much better and faster. If you are using linux or mac, then use the "zstd" command. In windows, check out https://mcmilk.de/projects/7-Zip-ZStd/ or other suitable alternatives.
Contact
If you have questions, please contact the corresponding author of the manuscript or post an issue in the GitHub issue tracker for this repository.