A developmental lineage-based gene co-expression network for mouse pancreatic beta-cells
Results from iterative refined WGCNA network analysis. The file includes 2 tables in separate tabs. 1) Gene Membership: full listing of the 15,949 expressed genes in the dataset and their module/meta-module membership. This table provides gene names, select gene functional classification annotation (T2D associated expression change, transcription factor, signaling molecule or lincRNA), gene population specific expression, gene meta-module and module assignment in the network, eigengene connectivity (kMe), gene expression profile, and gene expression values in each sample (normalized counts). 2) Modules: details on the 91 module detected by our analysis, including module size, module-density, and module eigengenes.
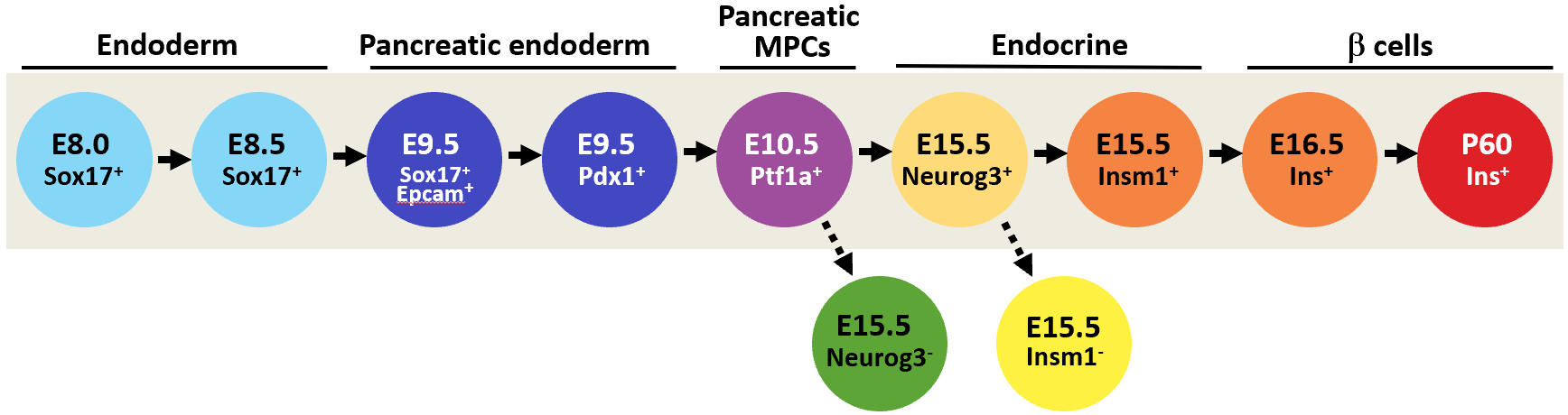
| Module Assignment | Endoderm | Pancreatic Endoderm | Pancreatic MPCs | Endocrine | Beta cells | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Metamodule | Module | Module_iWGCNA | # Genes | Eigengene Profile | Sox17 E8.0 | Sox17 E8.5 | Sox17 Epcam E9.5 | Pdx1 E9.5 | Ptf1a E10.5 | Neurog3 E15.5 KO | Neurog3 E15.5 | Insm1 E15.5 KO | Insm1 E15.5 | Ins E16.5 | Ins P60 |
| Metamodule | Module | Module_iWGCNA | # Genes | Eigengene Profile | Sox17 E8.0 | Sox17 E8.5 | Sox17 Epcam E9.5 | Pdx1 E9.5 | Ptf1a E10.5 | Neurog3 E15.5 KO | Neurog3 E15.5 | Insm1 E15.5 KO | Insm1 E15.5 | Ins E16.5 | Ins P60 |
Legend
- The data columns are averaged from the original eigengene values, per sample (N=3 per sample).